Gram-positive bacteria Computer Vision Project
Updated 2 years ago
Here are a few use cases for this project:
-
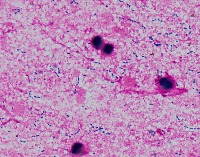
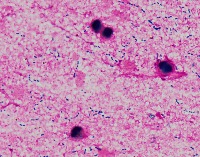
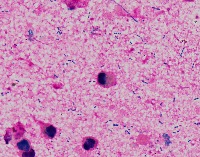
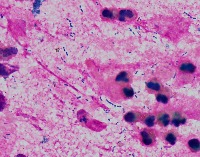
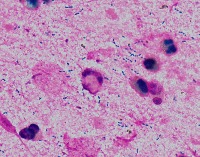
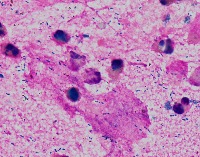
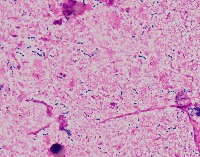
Medical Research: Researchers investigating the characteristics, growth patterns, and behaviors of Gram-positive bacteria, specifically Streptococcus pneumoniae (Str_pne), can use this computer vision model to identify and classify bacteria samples from microscopic images. This can help them in better understanding the pathogenesis and developing effective treatments.
-
Antibiotic Development: Pharmaceutical companies working on developing new antibiotics to fight Gram-positive bacterial infections can use this model to analyze the effects of potential drug candidates. They can perform comparative analysis of treated and untreated samples to assess the effectiveness of the drug in inhibiting the growth of the targeted bacteria.
-
Clinical Diagnostics: Diagnostic labs can employ this computer vision model to streamline the process of identifying Gram-positive bacteria in patient samples such as blood, sputum, and other body fluids. They can provide more timely and accurate diagnoses, thus facilitating appropriate treatment plans.
-
Antibiotic Resistance Monitoring: Public health organizations can use this model to track the prevalence of antibiotic-resistant strains of Gram-positive bacteria like Str_pne. By processing large amounts of bacterial samples from various sources, they can gain insights into the spread of antibiotic resistance and guide public health policies to combat this global health issue.
-
Environmental Monitoring: Environmental scientists can benefit from using this computer vision model to monitor the presence and concentrations of Gram-positive bacteria in different environmental samples, such as water and soil. This information can help evaluate microbial contamination levels and the impact of pollution on natural ecosystems.
Build Computer Vision Applications Faster with Supervision
Visualize and process your model results with our reusable computer vision tools.
Cite This Project
If you use this dataset in a research paper, please cite it using the following BibTeX:
@misc{
gram-positive-bacteria_dataset,
title = { Gram-positive bacteria Dataset },
type = { Open Source Dataset },
author = { terada shoma },
howpublished = { \url{ https://universe.roboflow.com/terada-shoma/gram-positive-bacteria } },
url = { https://universe.roboflow.com/terada-shoma/gram-positive-bacteria },
journal = { Roboflow Universe },
publisher = { Roboflow },
year = { 2022 },
month = { jul },
note = { visited on 2024-12-03 },
}