Arabidopsis Stomatal Counting Computer Vision Project
Updated 2 months ago
0
views0
downloadsMetrics
Introduction
Stomatal density analysis is essential for studying plant species but is traditionally time-consuming and labor-intensive when performed manually. Utilizing a deep learning model for stomatal counting can significantly reduce processing time and enable high-throughput analysis of Arabidopsis phenotypes. 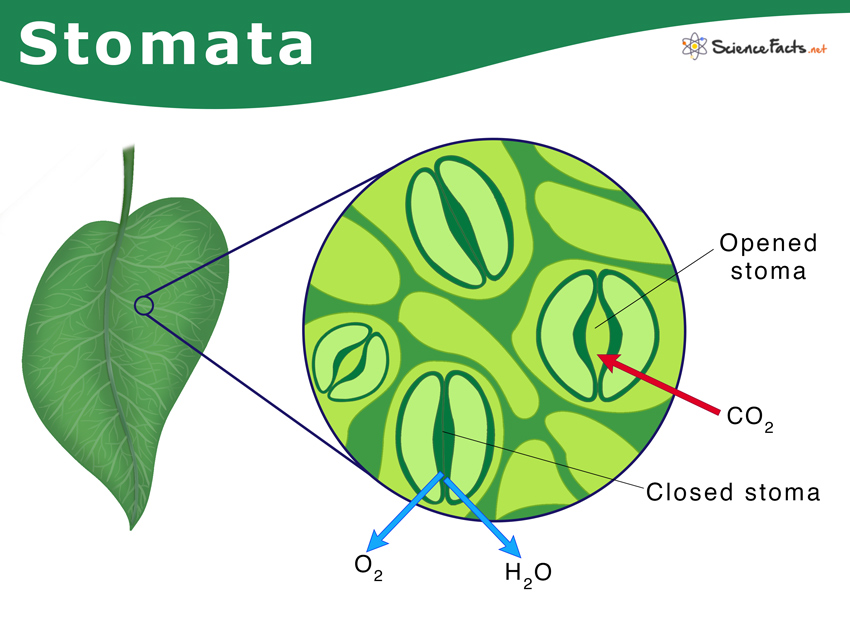 Figure 1. Illustration of stomata on the plant leaf surface.
Figure 1. Illustration of stomata on the plant leaf surface.
In this project, we created an annotated dataset using Arabidopsis leaf impressions and trained YOLO convolutional neural network (CNN) models for stomata detection. Currently, I am developing a pipeline to automatically count stomata from short videos of leaf impressions.
Dataset Preparation
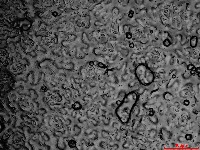
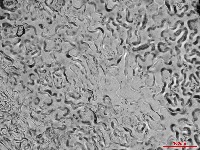
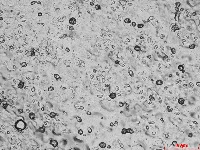
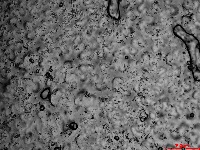
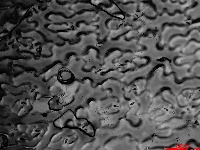
Microscopic images of leaf impressions are rarely fully in focus due to leaf surface curvature (Figure 2), complicating both manual and algorithm-based stomatal counting. To overcome this, videos with changing focal planes are used to capture the entire field of view.  Figure 2. Representative microscopic image of Arabidopsis leaf impression.
Figure 2. Representative microscopic image of Arabidopsis leaf impression.
However, CNNs cannot be trained directly on video data. Therefore, the dataset must consist of annotated images. I applied focus stacking technology to create fully focused images from multiple video frames (Figure 3). These images were then annotated to form the final dataset, available on the project page.  Figure 3. Pipeline for dataset preparation: focus stacking video frames to create a single image, followed by annotation using Roboflow.
Figure 3. Pipeline for dataset preparation: focus stacking video frames to create a single image, followed by annotation using Roboflow.
Training
YOLO CNN models were trained on this dataset. All versions of YOLOv8 models have been uploaded and can be accessed on this page. More advanced YOLOv9 and YOLOv10 models, which show improved performance, are still under development and will be integrated into the final project pipeline.
Implementation
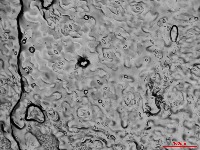
Converting videos to fully focused images is time-intensive, especially when processing large quantities. To address this, I am developing a method to apply the model directly to video data using a heatmap approach. The model detects each stoma across multiple frames, generating a heatmap where the numbers represent the frequency of stoma detection (Figure 4). This process is being automated with Python scripts to allow batch processing of multiple videos.  Figure 4. Heatmap generated by a script using the YOLOv9 CNN model: circles represent stomata, and numbers indicate the number of frames in which encircled stoma was detected.
Figure 4. Heatmap generated by a script using the YOLOv9 CNN model: circles represent stomata, and numbers indicate the number of frames in which encircled stoma was detected.
Use This Trained Model
Try it in your browser, or deploy via our Hosted Inference API and other deployment methods.
Build Computer Vision Applications Faster with Supervision
Visualize and process your model results with our reusable computer vision tools.
Cite This Project
If you use this dataset in a research paper, please cite it using the following BibTeX:
@misc{
arabidopsis-stomatal-counting_dataset,
title = { Arabidopsis Stomatal Counting Dataset },
type = { Open Source Dataset },
author = { Oleksii },
howpublished = { \url{ https://universe.roboflow.com/oleksii-hylbd/arabidopsis-stomatal-counting } },
url = { https://universe.roboflow.com/oleksii-hylbd/arabidopsis-stomatal-counting },
journal = { Roboflow Universe },
publisher = { Roboflow },
year = { 2024 },
month = { oct },
note = { visited on 2024-12-23 },
}