OnePetri Computer Vision Project
Updated 2 years ago
Metrics
Background Information
This dataset was created by Michael Shamash and contains the images used to train the OnePetri plaque detection model (plaque detection model v1.0).
In microbiology, a plaque is defined as a “clear area on an otherwise opaque field of bacteria that indicates the inhibition or dissolution of the bacterial cells by some agent, either a virus or an antibiotic. Plaques are a sensitive laboratory indicator of the presence of some anti-bacterial factor.” When working with bacteriophages (phages), viruses which can only infect and kill bacteria, scientists often need to perform the time-intensive monotonous task of counting plaques on Petri dishes. To help solve this problem I developed OnePetri, a set of machine learning models and a mobile phone application (currently iOS-only) that accelerates common microbiological Petri dish assays using AI.
A task that once took microbiologists several minutes to do per Petri dish (adds up quickly considering there are often tens of Petri dishes to analyze at a time!) could now be mostly automated thanks to computer vision, and completed in a matter of seconds.
App in Action
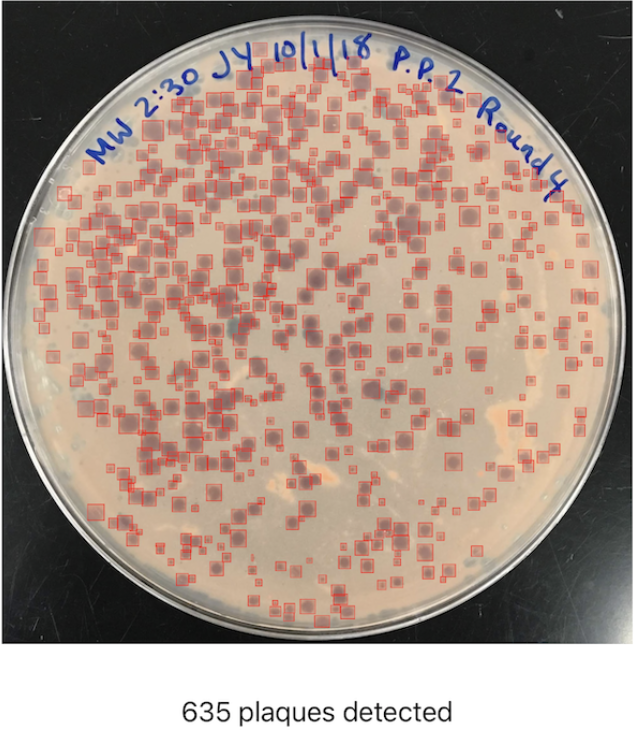
Example Image

A total of 43 source images were used in this dataset with the following split: 29 training, 9 validation, 5 testing (2505 images after preprocessing and augmentations are applied).
OnePetri is a mobile phone application (currently iOS-only) which accelerates common microbiological Petri dish assays using AI. OnePetri's YOLOv5s plaque detection model was trained on a diverse set of images from the HHMI's SEA-PHAGES program, many of which are included in this dataset. This project wouldn't be possible without their support!
The following pre-processing options were applied:
- Auto-orient
- Tile image into 5 rows x 5 columns
- Resize tiles to 416px x 416px
The following augmentation options were applied:
- Grayscale (35% of images)
- Hue shift (-45deg to +45deg)
- Blur up to 2px
- Mosaic
For more information and to download OnePetri please visit: https://onepetri.ai/.
Use This Trained Model
Try it in your browser, or deploy via our Hosted Inference API and other deployment methods.
Build Computer Vision Applications Faster with Supervision
Visualize and process your model results with our reusable computer vision tools.
Cite This Project
If you use this dataset in a research paper, please cite it using the following BibTeX:
@misc{
onepetri_dataset,
title = { OnePetri Dataset },
type = { Open Source Dataset },
author = { Michael Shamash RF Universe },
howpublished = { \url{ https://universe.roboflow.com/michael-shamash-rf-universe/onepetri } },
url = { https://universe.roboflow.com/michael-shamash-rf-universe/onepetri },
journal = { Roboflow Universe },
publisher = { Roboflow },
year = { 2022 },
month = { aug },
note = { visited on 2024-12-22 },
}